When we want to estimate parameters from data (e.g., from binding, kinetics, or electrophysiology experiments), there are two tasks: (i) estimate the most likely values, and (ii) equally importantly, estimate the uncertainty in those values. After all, if the uncertainty is huge, it’s hard to say we really know the parameters. We also need to choose the model in the first place, which is an extremely important task, but that is beyond the scope of this discussion.
Continue reading









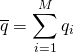 . We can also calculate the standard deviation σ of these values in the usual way as the square root of the variance. Both of these quantities will approach their “true” values (based on the simulation protocol) with enough sampling – with large enough
. We can also calculate the standard deviation σ of these values in the usual way as the square root of the variance. Both of these quantities will approach their “true” values (based on the simulation protocol) with enough sampling – with large enough